A spatial optimization algorithm blending Voronoi relaxation and 2-opt TSP refinement
Year : 2024
Author : Chan Yen Fen
Medium : JavaScript(P5JS); Python(Rhino Grasshopper)
Voronoi Relax TSP is a computational drawing system that transforms image data into a coherent, optimized path using spatial reasoning and classic algorithmic techniques. Starting with density-aware point sampling based on image brightness, it applies iterative Voronoi relaxation to refine point distribution. Once stabilized, the system constructs a path using the Nearest Neighbor heuristic and improves it with a customized 2-opt optimization guided by spatial queries via quadtree acceleration.
The result is a visually engaging and efficient traversal of space. For example, with 200 points, the algorithm reduces the path length by 15.2%, and with 1000 points, the reduction improves to 6.9%. This approach is ideal for generative art, pen plotting, and algorithmic design.



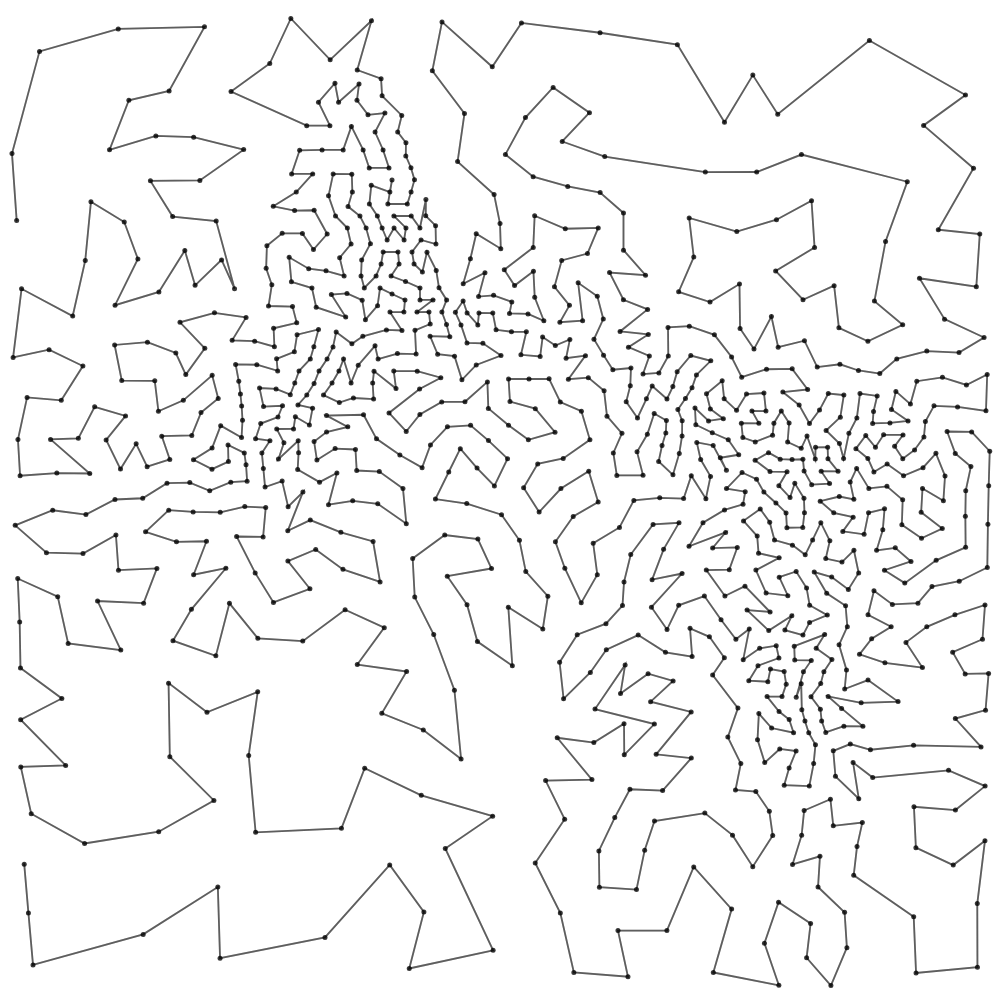
A sample web preview link is provided below, and you can view a 1000-point animation by clicking the link. To see the optimization metrics, click the button—note that the animation will begin once computation is complete(approx 10 seconds).
The TSP2opt algorithm is later implemented in Rhino Grasshopper using Python—available both as a standalone script and as custom components—since only a few existing plugins offer comparable functionality for sorting points into optimized traversal paths.
The optimizer aims to:
- Construct a trajectory that visits all points consecutively in the shortest possible path
- Guarantee that the resulting path has no self-intersections
Two Grasshopper variants are provided:
- TSP2opt_Manual – runs fast when parameters are tuned in advance, ideal as a baseline for small to medium datasets.
- TSP2opt_Adaptive – automatically estimates parameters from point density, more robust for large-scale point sets.
Sample code:
# Manual variant — user tunes the knobs
def tsp2opt_manual(points, max_iter=1000, radius=0.5):
path = nearest_neighbor(points) # seed
for _ in range(max_iter):
i, j = pick_segments(path, radius) # via spatial query / KDTree
if crossing(path[i], path[i+1], path[j], path[j+1]):
path = two_opt_swap(path, i, j) # 2-opt reversal
return path
# Adaptive variant — derives radius & iter from data
def tsp2opt_adaptive(points, mode="balanced"):
path = nearest_neighbor(points) # seed
# --- auto radius (matches your code idea) ---
L_med = median_edge_length(path) # from seed path
S_nn = median_nn_distance(points) # from KDTree
base = max(L_med, S_nn)
k = 3.0 if mode == "balanced" else (2.0 if mode == "fast" else 5.0)
radius = max(1e-6, k * base) # <- your current rule
# --- auto max_iter (matches your code exactly) ---
C0 = estimate_crossings(path, radius) # via midpoint KDTree
n = len(path)
max_iter = min(300, 10 + int(0.5 * n) + 2 * C0) # <- your current cap
# --- optimization loop ---
for _ in range(max_iter):
i, j = pick_segments(path, radius)
if crossing(path[i], path[i+1], path[j], path[j+1]):
path = two_opt_swap(path, i, j)
return pathTo showcase performance, the GitHub repository includes benchmarks with 1k, 5k, and 10k points.